PUREfrex® is a cell-free protein synthesis system in which the transcription of template DNA into mRNA is coupled to the translation of that mRNA into protein. Termination is occured when a stop codon is recogonized by release factors (RF1 and RF2) and the polypeptide is released.
Bacterial release factors include RF1, RF2, and RF3. RF1 recognizes UAA and UAG while RF2 recognizes UAA and UGA. PUREfrex® includes RF1 and RF2, therefore all three of the different stop codons can be used.
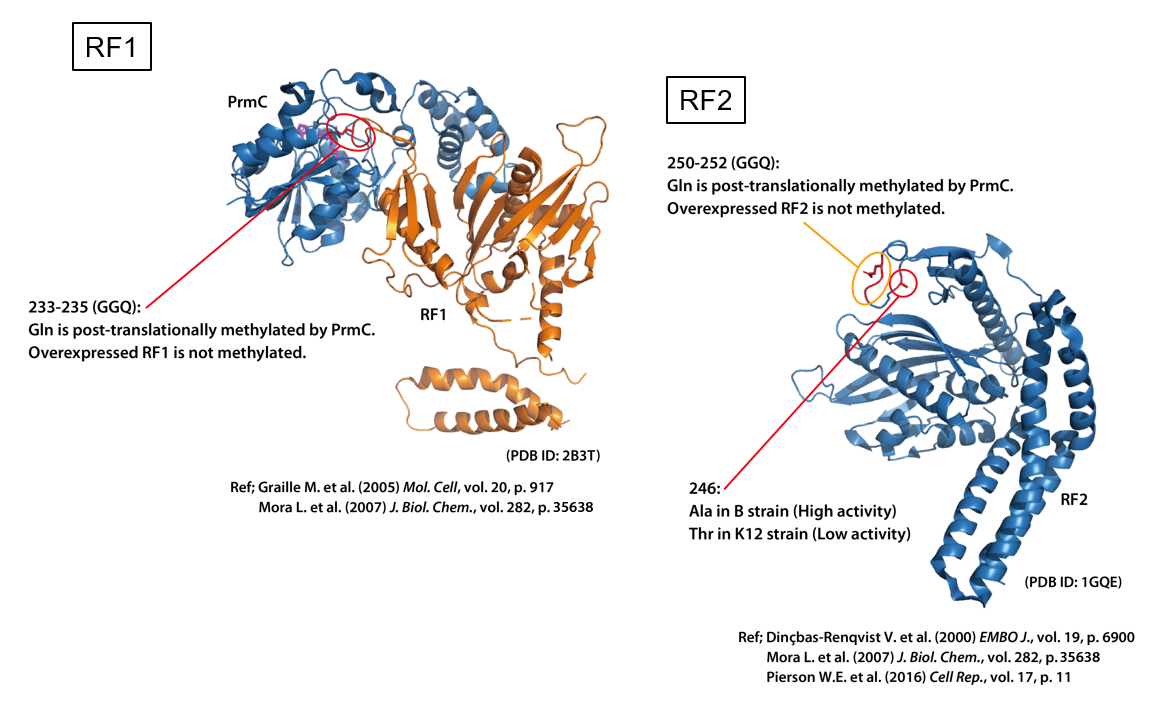
1. Methylation of release factors RF1 and RF2
Bacterial release factors RF1 and RF2 are methylated on the Gln residue of a universally conserved tripeptide motif GGQ. In vitro experiments have shown that the activity of RF1 and RF2 is stimulated by Gln methylation.
Here, we demonstrated that the comparison of activity of RF1 and RF2 with and without methylation by using translational readthrough.
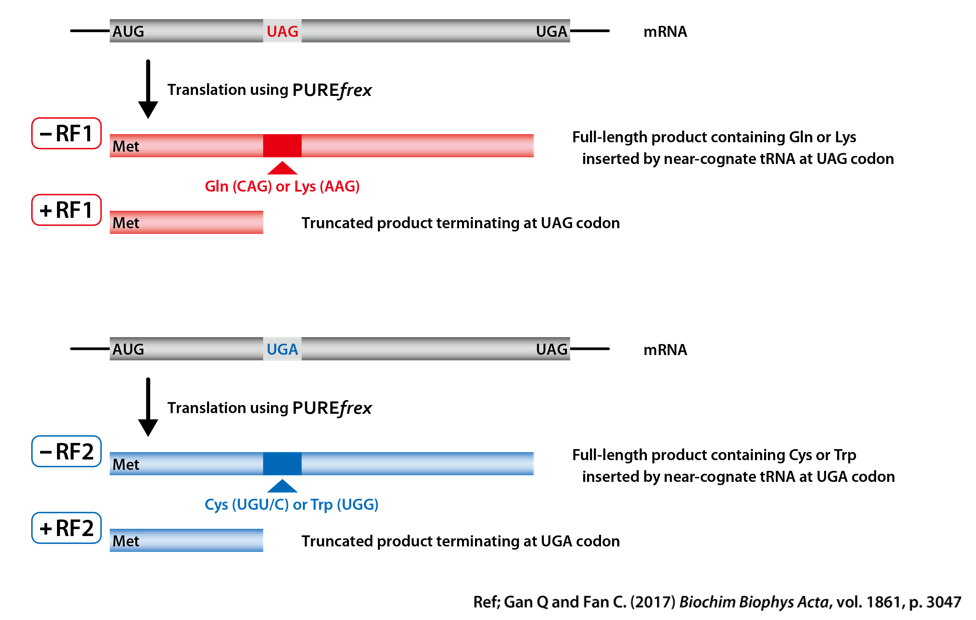
2. Preparation of template DNA for translational readthrough
2-1. Design of template DNA
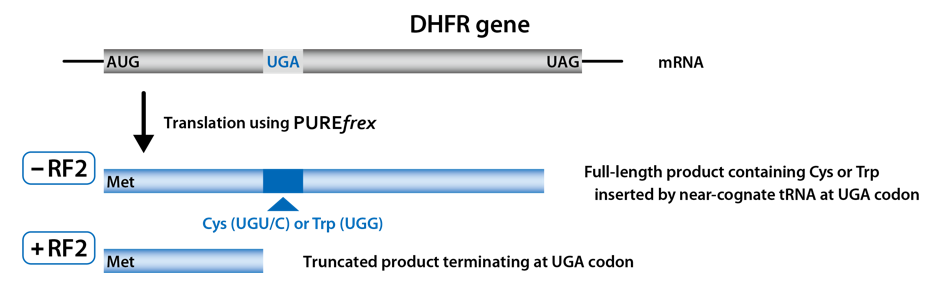
DHFR (dihydrofolate reductase) was used as a model protein.
Two different stop codons were inserted in the ORF.
The first one was in the middle of DHFR gene and the second one was at the end of DHFR gene.
| Codon | Release factors | |
|---|---|---|
| DNA | RNA | |
| TAG | UAG | RF1 |
| TAA | UAA | RF1, RF2 |
| TGA | UGA | RF2 |
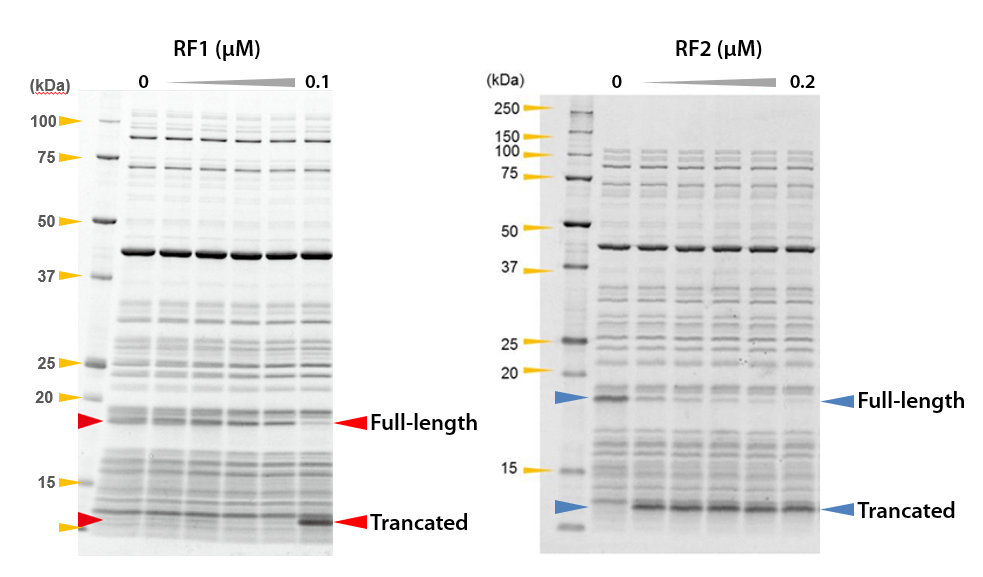
2-2. Results of translational readthrough
2-2-1. In case of RF1
DHFR was synthesized by preparing reaction mixture with different final concentrations of RF1. The results are shown on the left side of figure.
Full-length DHFR was synthesized in the reaction mixture without RF1 (0 µM), and trancated DHFR was synthesized in the reaction mixture with RF1. It was confirmed that translational readthrough occurred in the absence of RF1 or with a low concentration of RF1.
2-2-2. In case of RF2
DHFR was synthesized by preparing reaction mixture with different final concentrations of RF2. The results are shown on the right side of figure.
Full-length DHFR was synthesized in the reaction mixture without RF2 (0 µM), and trancated DHFR was synthesized in the reaction mixture with RF2. It was confirmed that translational readthrough occurred in the absence of RF2.
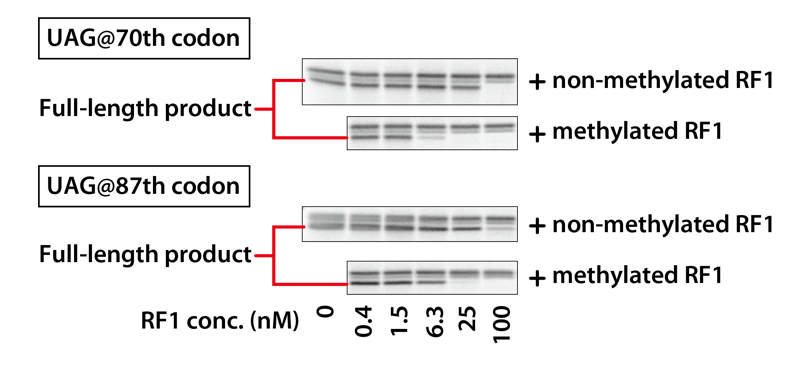
3. Comparison of RF1 activity
3-1. Materials and Methods
Reaction mixtures with different final concentrations of RF1 were prepared, and the size and amount of DHFR synthesized in that reaction mixtures were compared.
3-1-1. Two Template DNAs
– UAG @70th codon
UAG stop codon is present at the 70th position and UGA stop codon terminate the translation with RF2.
– UAG @87th codon
UAG stop codon is present at the 87th position and UGA stop codon terminate the translation with RF2.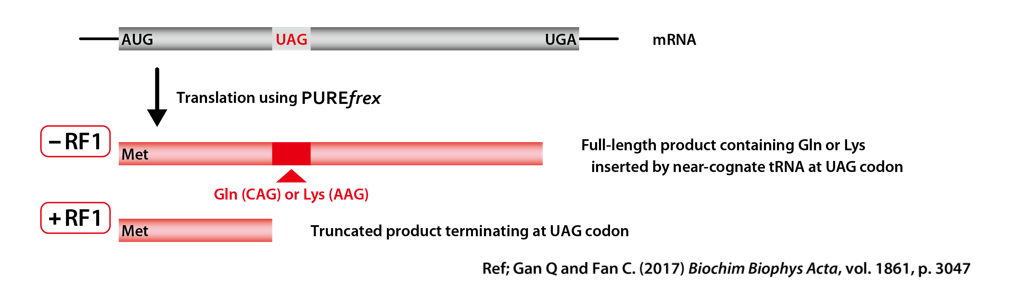
3-1-2. RF1 preparation
– Methylated RF1
– Non-methylated RF1
3-1-3. Reaction mixture
– Customized PUREfrex® without RF1
3-2. Results
The synthesized DHFRs were compared of SDS-PAGE protein patterns. In the result, only the full-length part is shown.
Methylated RF1 suppressed readthrough more efficiently than non-methylated RF1, as full-length DHFR was undetectable at lower concentrations than non-methylated RF1. Methylated RF1 has a higher specific activity than non-methylated RF1.
4. Comparison of RF2 activity
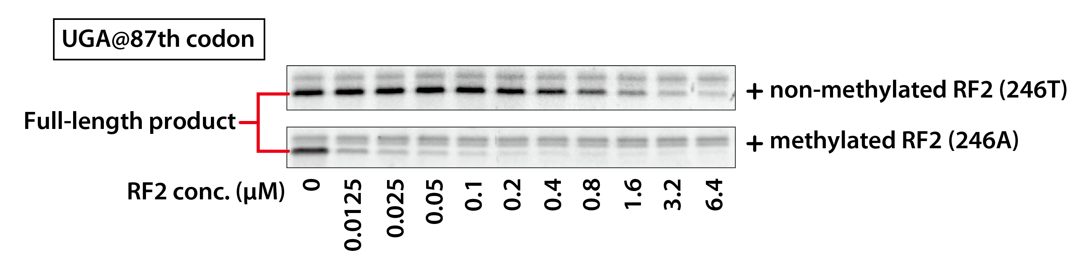
4-1. Materials and Methods
Reaction mixtures with different final concentrations of RF2 were prepared, and the size and amount of DHFR synthesized in that reaction mixtures were compared.
4-1-1. Template DNA
– UGA @87th codon
UGA stop codon is present at the 87th position and UAG stop codon terminate the translation with RF1.
4-1-2. RF2 preparation
– Methylated RF2 (246A): Ala residue at position 246
– Non-methylated RF2 (246T) : Thr residue at position 246
Escherichia coli K12 strains are partially deficient in RF2 activity due to the presence of a Thr residue at position 246 instead of Ala.
4-1-3. Reaction mixture
– Customized PUREfrex® without RF2
4-2. Results
The synthesized DHFRs were compared of SDS-PAGE protein patterns. In the result, only the full-length part is shown.
Methylated RF2 (246A) suppressed readthrough more efficiently than non-methylated RF2 (246T), as full-length DHFR was undetectable at lower concentrations than non-methylated RF2 (246T). Methylated RF2 (246A) has a higher specific activity than non-methylated RF2 (246T).
5. Conclusion
Methylated RF1 and RF2 suppressed readthrough more efficiently than non-methylated RF1 and RF2.
For proteins that had low amounts of synthesized products due to incomplete termination of translation by readthrough, methylated RF1 and RF2 are expected to improve polypeptide release, resulting in higher amounts of synthesized proteins.

